Note
Go to the end to download the full example code.
Random Sampling in a Volume#
This example inspired by the discussion in
this PyVista discussion.
Using pyransame makes the solution easier and handles more complex scenarios.
This example is related to post-processing CFD data with a Eulerian description
of particle density.
import numpy as np
import pyvista as pv
import pyransame
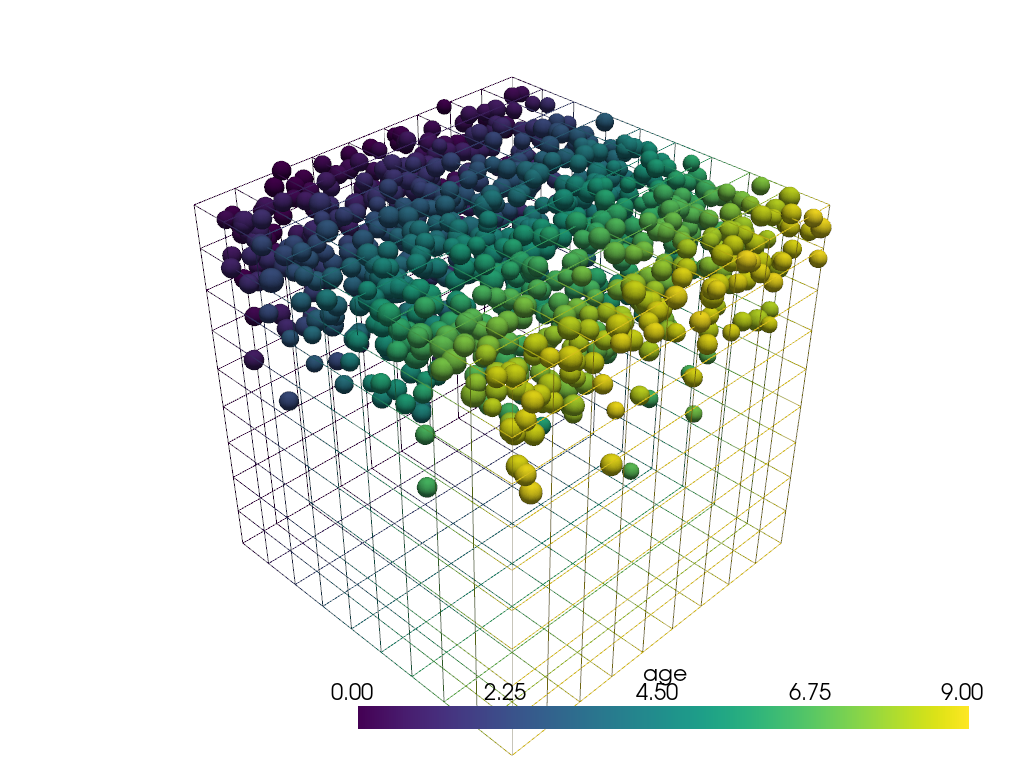
This example shows how pyransame can be used to sampled from datasets
that have volume. There is a cube of dimension 10 m x 10 m x 10 m
with particles described by a volume fraction in each cell.
In this synthetic dataset, the
particles tend to float and are located near the top (z-position).
cell_centers is used as cell_data is required for weighted sampling.
mesh = pv.ImageData(dimensions=(10, 10, 10))
mesh.cell_data["volume_frac"] = np.exp(mesh.cell_centers().points[:, 2])
mesh.cell_data["volume_frac"] /= np.sum(mesh["volume_frac"])
The particles have an age related to some history of the particle motion, in this case related to the y-position. Cell data is not required here as the point data can be interpolated onto the sampled points.
mesh["age"] = mesh.points[:, 1]
Sample the points with respect to the volume fraction to obtain a realistic particle distribution in the domain.
points = pyransame.random_volume_dataset(mesh, 1000, weights="volume_frac")
In this example, we also know the particles diameters come from a normal distribution that does not depend on the other particle attributes.
diameters = 0.03 * np.random.randn(1000) + 0.4
points["diameter"] = diameters
Plot.
cpos = [[32.0, 16.0, 10.0], [5.0, 3.9, 3.6], [-0.21, -0.076, 0.97]]
pl = pv.Plotter()
pl.add_mesh(mesh, style="wireframe")
spheres = points.glyph(geom=pv.Sphere(), scale="diameter", orient=False)
pl.add_mesh(spheres, scalars="age")
pl.show()
Total running time of the script: (0 minutes 7.930 seconds)